language:
- en
- zh
license: apache-2.0
tags:
- fine tune
- creative
- creative writing
- fiction writing
- plot generation
- sub-plot generation
- fiction writing
- story generation
- scene continue
- storytelling
- fiction story
- science fiction
- romance
- all genres
- story
- writing
- vivid prosing
- vivid writing
- fiction
- roleplaying
- bfloat16
- all use cases
- unsloth
- heretic
- uncensored
- abliterated
library_name: transformers
pipeline_tag: image-text-to-text
base_model:
- DavidAU/Qwen3.5-9B-Claude-4.6-OS-Auto-Variable-HERETIC-UNCENSORED-THINKING
<h2>Qwen3.5-9B-Claude-4.6-OS-Auto-Variable-HERETIC-UNCENSORED-THINKING-MAX-NEOCODE-IMATRIX-GGUF</h2>
<img src="valhalla.webp" style="float:right; width:300px; height:300px; padding:10px;">
Fine tune via Unsloth of Qwen 3.5 9B dense model using Claude-4.6-OS dataset (4 Claude datasets) on local hardware. Every attempt was made to ensure the training was "mild" and did not negatively affect the model's already incrediblely strong benchmarks.
Training on 4 datasets improves reasoning, and output generation and exceeds the root model on all benchmarks too.
This is also a HERETIC model, trained post "Heretic'ing" -> this model does what you want, no questions asked.
Fully uncensored.
Example generation(s) at the bottom of this page.
GGUF POWER UPS:
- DI-MATRIX (duel imatrix) of NEO and NEO-CODE imatrix datasets (by DavidAU).
- All Unsloth tensor enhancements.
- BF16 output tensor to improve both thinking and output.
- Custom Jinja template (by DavidAU/Nightmedia) to address some issues with the Qwen 3.5 model series (stablize and reduce overthinking).
A radically stronger, more potent GGUF for all use cases.
VISION:
- Vision (images) tested -> working with new training.
- You need an "mmproj" (just one) of these downloaded too, and placed in the same folder as the GGUF for images.
BENCHMARKS:
arc arc/e boolq hswag obkqa piqa wino
HERETIC version (this model):
mxfp8 0.425,0.521,0.791,0.664,0.374,0.744,0.645
HERETIC version (instruct):
mxfp8 0.624,0.820,0.886,0.663,0.442,0.763,0.681
Qwen3.5-9B (thinking)
mxfp8 0.417,0.458,0.623,0.634,0.338,0.737,0.639
Instruct version (as noted above in "benchmarks") is here:
https://huggingface.co/DavidAU/Qwen3.5-9B-Claude-4.6-OS-HERETIC-UNCENSORED-INSTRUCT
DE-CENSORING:
Performance
KLD of less than 1 is excellent, zero is perfect.
| Metric | This model | Original model (Qwen/Qwen3.5-9B) |
| :----- | :--------: | :---------------------------: |
| KL divergence | 0.0793 | 0 (by definition) |
| Refusals | 6/100 | 100/100 |
NOTES:
- Suggest min q4ks (non-imatrix) or IQ3S (imatrix).
- Tested with rep pen of 1 (off).
- Context: 256k (default).
IMPORTANT:
- Other versions in testing.
- Information from Qwen's repo below.
- Video portions of the model were NOT TESTED.
<B>Using an "uncensored" (refusals removed) model VS trained "uncensored" model</B>
Usually when you a tell a model to generate horror, swear or x-rated content this is all you have to do to get said content type.
In the case of this model, it will not refuse your request, however it needs to be "pushed" a bit / directed a bit more in SOME CASES.
Although this model will generated x-rated content too, likewise you need to tell it to use "slang" (and include the terms you want)
to get it generate the content correctly as the "expected" content level too.
Without these added directive(s), the content can be "bland" by comparison to an "uncensored model" or model trained on uncensored content.
Roughly, the model tries to generate the content but the "default" setting(s) are so "tame" it needs a push to generate at expected graphic,
cursing or explicit levels.
Even with minimal direction (ie, use these words to swear: x,y,z), this will be enough to push the model to generate the requested content in the ahh... expected format.
<B>Settings: CHAT / ROLEPLAY and/or SMOOTHER operation of this model:</B>
In "KoboldCpp" or "oobabooga/text-generation-webui" or "Silly Tavern" ;
Set the "Smoothing_factor" to 1.5
: in KoboldCpp -> Settings->Samplers->Advanced-> "Smooth_F"
: in text-generation-webui -> parameters -> lower right.
: In Silly Tavern this is called: "Smoothing"
NOTE: For "text-generation-webui"
-> if using GGUFs you need to use "llama_HF" (which involves downloading some config files from the SOURCE version of this model)
Source versions (and config files) of my models are here:
https://huggingface.co/collections/DavidAU/d-au-source-files-for-gguf-exl2-awq-gptq-hqq-etc-etc-66b55cb8ba25f914cbf210be
OTHER OPTIONS:
-
Increase rep pen to 1.1 to 1.15 (you don't need to do this if you use "smoothing_factor")
-
If the interface/program you are using to run AI MODELS supports "Quadratic Sampling" ("smoothing") just make the adjustment as noted.
<B>Highest Quality Settings / Optimal Operation Guide / Parameters and Samplers</B>
This a "Class 1" model:
For all settings used for this model (including specifics for its "class"), including example generation(s) and for advanced settings guide (which many times addresses any model issue(s)), including methods to improve model performance for all use case(s) as well as chat, roleplay and other use case(s) please see:
[ https://huggingface.co/DavidAU/Maximizing-Model-Performance-All-Quants-Types-And-Full-Precision-by-Samplers_Parameters ]
You can see all parameters used for generation, in addition to advanced parameters and samplers to get the most out of this model here:
[ https://huggingface.co/DavidAU/Maximizing-Model-Performance-All-Quants-Types-And-Full-Precision-by-Samplers_Parameters ]
Qwen3.5-9B
<img width="400px" src="https://qianwen-res.oss-accelerate.aliyuncs.com/logo_qwen3.5.png">

[!Note]
This repository contains model weights and configuration files for the post-trained model in the Hugging Face Transformers format.
These artifacts are compatible with Hugging Face Transformers, vLLM, SGLang, KTransformers, etc.
Over recent months, we have intensified our focus on developing foundation models that deliver exceptional utility and performance. Qwen3.5 represents a significant leap forward, integrating breakthroughs in multimodal learning, architectural efficiency, reinforcement learning scale, and global accessibility to empower developers and enterprises with unprecedented capability and efficiency.
Qwen3.5 Highlights
Qwen3.5 features the following enhancement:
-
Unified Vision-Language Foundation: Early fusion training on multimodal tokens achieves cross-generational parity with Qwen3 and outperforms Qwen3-VL models across reasoning, coding, agents, and visual understanding benchmarks.
-
Efficient Hybrid Architecture: Gated Delta Networks combined with sparse Mixture-of-Experts deliver high-throughput inference with minimal latency and cost overhead.
-
Scalable RL Generalization: Reinforcement learning scaled across million-agent environments with progressively complex task distributions for robust real-world adaptability.
-
Global Linguistic Coverage: Expanded support to 201 languages and dialects, enabling inclusive, worldwide deployment with nuanced cultural and regional understanding.
-
Next-Generation Training Infrastructure: Near-100% multimodal training efficiency compared to text-only training and asynchronous RL frameworks supporting massive-scale agent scaffolds and environment orchestration.
For more details, please refer to our blog post Qwen3.5.
Model Overview
- Type: Causal Language Model with Vision Encoder
- Training Stage: Pre-training & Post-training
- Language Model
- Number of Parameters: 9B
- Hidden Dimension: 4096
- Token Embedding: 248320 (Padded)
- Number of Layers: 32
- Hidden Layout: 8 × (3 × (Gated DeltaNet → FFN) → 1 × (Gated Attention → FFN))
- Gated DeltaNet:
- Number of Linear Attention Heads: 32 for V and 16 for QK
- Head Dimension: 128
- Gated Attention:
- Number of Attention Heads: 16 for Q and 4 for KV
- Head Dimension: 256
- Rotary Position Embedding Dimension: 64
- Feed Forward Network:
- Intermediate Dimension: 12288
- LM Output: 248320 (Padded)
- MTP: trained with multi-steps
- Context Length: 262,144 natively and extensible up to 1,010,000 tokens.
Benchmark Results
Language
<div style="font-family:-apple-system,BlinkMacSystemFont,'Segoe UI',Roboto,sans-serif;max-width:1000px;margin:0 auto;padding:16px 0">
<table style="width:100%;border-collapse:collapse;font-size:13px">
<thead><tr>
<th style="padding:10px 7px;text-align:left;font-weight:600;border-bottom:2px solid #7c3aed;color:#7c3aed"></th><th style="padding:10px 7px;text-align:center;font-weight:500;border-bottom:2px solid #7c3aed;color:#7c3aed;font-size: 14px;">GPT-OSS-120B</th><th style="padding:10px 7px;text-align:center;font-weight:500;border-bottom:2px solid #7c3aed;color:#7c3aed;font-size: 14px;">GPT-OSS-20B</th><th style="padding:10px 7px;text-align:center;font-weight:500;border-bottom:2px solid #7c3aed;color:#7c3aed;font-size: 14px;">Qwen3-Next-80B-A3B-Thinking</th><th style="padding:10px 7px;text-align:center;font-weight:500;border-bottom:2px solid #7c3aed;color:#7c3aed;font-size: 14px;">Qwen3-30BA3B-Thinking-2507</th><th style="padding:10px 7px;text-align:center;font-weight:500;border-bottom:2px solid #7c3aed;color:#7c3aed;font-size: 14px;">Qwen3.5-9B</th><th style="padding:10px 7px;text-align:center;font-weight:500;border-bottom:2px solid #7c3aed;color:#7c3aed;font-size: 14px;">Qwen3.5-4B</th></tr></thead>
<tbody>
<tr><td colspan="7" style="padding:8px 12px;font-weight:600;color:#7c3aed;border-bottom:1px solid rgba(124, 58, 237, 0.2);background:rgba(124, 58, 237, 0.1)">Knowledge & STEM</td></tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">MMLU-Pro</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">80.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">74.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">82.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">80.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">82.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">79.1</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">MMLU-Redux</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">91.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">87.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">92.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">91.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">91.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">88.8</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">C-Eval</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">76.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">71.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">89.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">87.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">88.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">85.1</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">SuperGPQA</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">54.6</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">48.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">60.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">56.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">58.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">52.9</td>
</tr>
<tr>
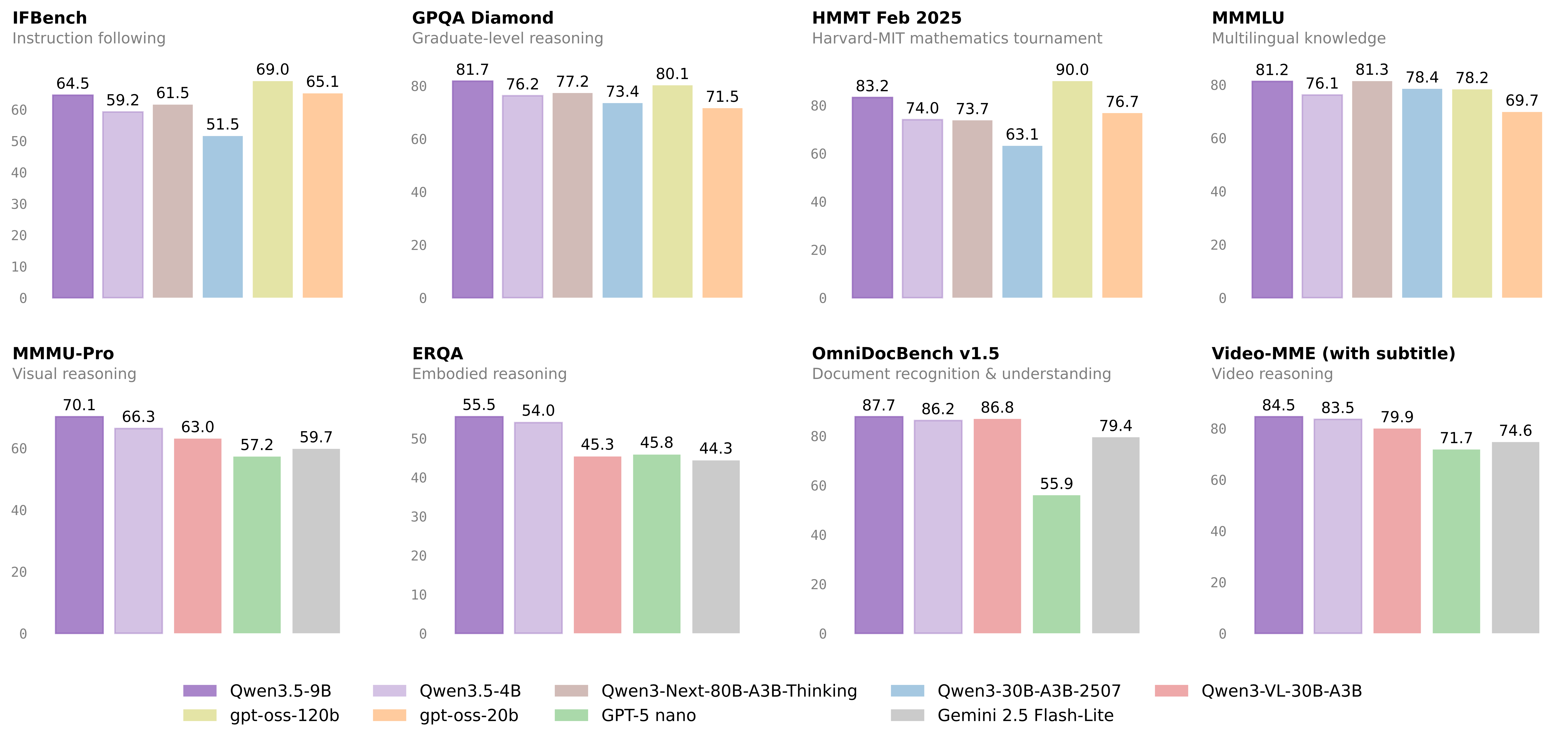
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">GPQA Diamond</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">80.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">71.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">77.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">73.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">81.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">76.2</td>
</tr>
<tr><td colspan="7" style="padding:8px 12px;font-weight:600;color:#7c3aed;border-bottom:1px solid rgba(124, 58, 237, 0.2);background:rgba(124, 58, 237, 0.1)">Instruction Following</td></tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">IFEval</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">88.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">88.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">88.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">88.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">91.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">89.8</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">IFBench</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">69.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">65.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">61.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">51.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">64.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">59.2</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">MultiChallenge</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">45.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">40.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">51.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">46.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">54.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">49.0</td>
</tr>
<tr><td colspan="7" style="padding:8px 12px;font-weight:600;color:#7c3aed;border-bottom:1px solid rgba(124, 58, 237, 0.2);background:rgba(124, 58, 237, 0.1)">Long Context</td></tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">AA-LCR</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">50.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">30.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">51.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">49.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">63.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">57.0</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">LongBench v2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">48.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">45.6</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">48.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">44.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">55.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">50.0</td>
</tr>
<tr><td colspan="7" style="padding:8px 12px;font-weight:600;color:#7c3aed;border-bottom:1px solid rgba(124, 58, 237, 0.2);background:rgba(124, 58, 237, 0.1)">Reasoning & Coding</td></tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">HMMT Feb 25</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">90.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">76.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">73.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">63.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">83.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">74.0</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">HMMT Nov 25</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">90.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">81.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">81.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">73.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">82.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">76.8</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">LiveCodeBench v6</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">82.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">74.6</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">68.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">66.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">65.6</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">55.8</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">OJBench</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">41.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">36.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">29.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">25.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">29.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">24.1</td>
</tr>
<tr><td colspan="7" style="padding:8px 12px;font-weight:600;color:#7c3aed;border-bottom:1px solid rgba(124, 58, 237, 0.2);background:rgba(124, 58, 237, 0.1)">General Agent</td></tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">BFCL-V4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">49.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">42.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">66.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">50.3</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">TAU2-Bench</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">57.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">41.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">79.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">79.9</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">VITA-Bench</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">29.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">14.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">29.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">22.0</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">DeepPlanning</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">0.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">4.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">18.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">17.6</td>
</tr>
<tr><td colspan="7" style="padding:8px 12px;font-weight:600;color:#7c3aed;border-bottom:1px solid rgba(124, 58, 237, 0.2);background:rgba(124, 58, 237, 0.1)">Multilingualism</td></tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">MMMLU</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">78.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">69.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">81.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">78.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">81.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">76.1</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">MMLU-ProX</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">74.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">67.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">73.6</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">69.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">76.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">71.5</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">NOVA-63</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">51.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">48.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">53.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">52.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">55.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">54.3</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">INCLUDE</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">74.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">65.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">78.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">74.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">75.6</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">71.0</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">Global PIQA</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">84.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">79.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">83.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">80.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">83.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">78.9</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">PolyMATH</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">54.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">30.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">62.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">52.6</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">57.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">51.1</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">WMT24++</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">74.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">67.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">57.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">69.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">72.6</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">66.6</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">MAXIFE</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">83.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">80.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">79.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">77.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">83.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">78.0</td>
</tr>
</tbody>
</table>
<p style="margin-top:12px;font-size:11px;opacity:0.7">
* TAU2-Bench: we follow the official setup except for the airline domain, where all models are evaluated by applying the fixes proposed in the Claude Opus 4.5 system card.<br>
<br>
* MMLU-ProX: we report the averaged accuracy on 29 languages.<br>
* WMT24++: a harder subset of WMT24 after difficulty labeling and rebalancing; we report the averaged scores on 55 languages using XCOMET-XXL.<br>
* MAXIFE: we report the accuracy on English + multilingual original prompts (totally 23 settings).<br>
* Empty cells (--) indicate scores not yet available or not applicable.
</p>
</div>
Vision Language
<div style="font-family:-apple-system,BlinkMacSystemFont,'Segoe UI',Roboto,sans-serif;max-width:1000px;margin:0 auto;padding:16px 0">
<table style="width:100%;border-collapse:collapse;font-size:13px">
<thead><tr>
<th style="padding:10px 7px;text-align:left;font-weight:600;border-bottom:2px solid #7c3aed;color:#7c3aed"></th><th style="padding:10px 7px;text-align:center;font-weight:500;border-bottom:2px solid #7c3aed;color:#7c3aed;font-size: 14px;">GPT-5-Nano-2025-08-07</th><th style="padding:10px 7px;text-align:center;font-weight:500;border-bottom:2px solid #7c3aed;color:#7c3aed;font-size: 14px;">Gemini-2.5-Flash-Lite</th><th style="padding:10px 7px;text-align:center;font-weight:500;border-bottom:2px solid #7c3aed;color:#7c3aed;font-size: 14px;">Qwen3-VL-30B-A3B</th><th style="padding:10px 7px;text-align:center;font-weight:500;border-bottom:2px solid #7c3aed;color:#7c3aed;font-size: 14px;">Qwen3.5-9B</th><th style="padding:10px 7px;text-align:center;font-weight:500;border-bottom:2px solid #7c3aed;color:#7c3aed;font-size: 14px;">Qwen3.5-4B</th></tr></thead>
<tbody>
<tr><td colspan="6" style="padding:8px 12px;font-weight:600;color:#7c3aed;border-bottom:1px solid rgba(124, 58, 237, 0.2);background:rgba(124, 58, 237, 0.1)">STEM and Puzzle </td></tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">MMMU</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">75.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">73.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">76.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">78.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">77.6</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">MMMU-Pro</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">57.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">59.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">63.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">70.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">66.3</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">MathVision</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">62.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">52.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">65.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">78.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">74.6</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">Mathvista(mini)</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">71.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">72.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">81.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">85.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">85.1</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">We-Math</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">62.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">32.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">70.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">75.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">75.4</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">DynaMath</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">78.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">69.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">80.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">83.6</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">83.3</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">ZEROBench</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">1.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">1.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">0.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">3.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">3.0</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">ZEROBench_sub</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">22.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">19.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">23.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">31.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">26.3</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">VlmsAreBlind</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">66.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">68.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">72.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">93.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">92.6</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">BabyVision</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">14.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">17.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">18.6</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">28.6/25.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">16.0/19.1</td>
</tr>
<tr><td colspan="6" style="padding:8px 12px;font-weight:600;color:#7c3aed;border-bottom:1px solid rgba(124, 58, 237, 0.2);background:rgba(124, 58, 237, 0.1)">General VQA</td></tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">RealWorldQA</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">71.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">72.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">77.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">80.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">79.5</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">MMStar</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">68.6</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">69.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">75.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">79.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">78.3</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">MMBench<sub><small>EN-DEV-v1.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">80.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">82.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">88.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">90.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">89.4</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">SimpleVQA</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">46.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">54.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">54.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">51.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">43.4</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">HallusionBench</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">58.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">64.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">66.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">69.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">65.0</td>
</tr>
<tr><td colspan="6" style="padding:8px 12px;font-weight:600;color:#7c3aed;border-bottom:1px solid rgba(124, 58, 237, 0.2);background:rgba(124, 58, 237, 0.1)">Text Recognition and Document Understanding</td></tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">OmniDocBench1.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">55.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">79.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">86.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">87.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">86.2</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">CharXiv(RQ)</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">50.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">56.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">56.6</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">73.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">70.8</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">MMLongBench-Doc</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">31.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">46.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">47.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">57.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">54.2</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">CC-OCR</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">58.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">72.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">77.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">79.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">76.7</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">AI2D_TEST</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">81.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">85.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">86.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">90.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">89.6</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">OCRBench</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">75.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">82.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">83.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">89.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">85.0</td>
</tr>
<tr><td colspan="6" style="padding:8px 12px;font-weight:600;color:#7c3aed;border-bottom:1px solid rgba(124, 58, 237, 0.2);background:rgba(124, 58, 237, 0.1)">Spatial Intelligence</td></tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">ERQA</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">45.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">44.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">45.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">55.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">54.0</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">CountBench</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">80.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">79.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">90.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">97.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">96.3</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">RefCOCO(avg)</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">89.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">89.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">88.1</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">EmbSpatialBench</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">74.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">66.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">80.6</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">83.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">81.3</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">RefSpatialBench</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">12.6</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">11.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">54.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">58.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">54.6</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">LingoQA</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">57.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">17.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">62.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">80.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">74.4</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">Hypersim</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">11.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">13.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">12.5</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">Nuscene</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">10.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">11.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">9.9</td>
</tr>
<tr><td colspan="6" style="padding:8px 12px;font-weight:600;color:#7c3aed;border-bottom:1px solid rgba(124, 58, 237, 0.2);background:rgba(124, 58, 237, 0.1)">Video Understanding</td></tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">VideoMME<sub><small>(w sub.)</sub></small></td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">71.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">74.6</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">79.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">84.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">83.5</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">VideoMME<sub><small>(w/o sub.)</sub></small></td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">66.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">72.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">73.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">78.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">76.9</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">VideoMMMU</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">63.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">69.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">75.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">78.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">74.1</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">MLVU</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">69.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">78.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">78.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">84.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">82.8</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">MVBench</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">72.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">74.4</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">71.2</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">LVBench</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">60.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">59.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">70.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">66.4</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">MMVU</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">63.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">65.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">66.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">67.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">64.9</td>
</tr>
<tr><td colspan="6" style="padding:8px 12px;font-weight:600;color:#7c3aed;border-bottom:1px solid rgba(124, 58, 237, 0.2);background:rgba(124, 58, 237, 0.1)">Visual Agent </td></tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">ScreenSpot Pro</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">60.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">65.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">60.3</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">OSWorld-Verified</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">30.6</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">41.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">35.6</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">AndroidWorld</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">--</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">55.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">57.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">58.6</td>
</tr>
<tr><td colspan="6" style="padding:8px 12px;font-weight:600;color:#7c3aed;border-bottom:1px solid rgba(124, 58, 237, 0.2);background:rgba(124, 58, 237, 0.1)">Tool Calling</td></tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">TIR-Bench</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">18.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">21.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">22.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">45.6/31.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">38.9/29.9</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">V*</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">68.1</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">69.6</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">83.2</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">90.1/88.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">84.3/86.4</td>
</tr>
<tr><td colspan="6" style="padding:8px 12px;font-weight:600;color:#7c3aed;border-bottom:1px solid rgba(124, 58, 237, 0.2);background:rgba(124, 58, 237, 0.1)">Medical VQA</td></tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">SLAKE</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">57.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">65.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">68.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">79.0</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">76.1</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">PMC-VQA</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">37.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">48.8</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">51.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">57.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">55.5</td>
</tr>
<tr>
<td style="padding:7px 7px;padding-left:20px;border-bottom:1px solid rgba(128, 128, 128, 0.15);">MedXpertQA-MM</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">26.7</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">35.3</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">35.5</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">49.9</td>
<td style="padding:7px 7px;text-align:center;border-bottom:1px solid rgba(128, 128, 128, 0.15)">42.9</td>
</tr>
</tbody>
</table>
<p style="margin-top:12px;font-size:11px;opacity:0.7">
* MathVision: our model’s score is evaluated using a fixed prompt, e.g., “Please reason step by step, and put your final answer within \boxed{}.” For other models, we report the higher score between runs with and without the \boxed{} formatting.<br>
* BabyVision: scores reported as "with CI / without CI".<br>
* TIR-Bench and V*: scores reported as "with CI / without CI".<br>
* Empty cells (--) indicate scores not yet available or not applicable.
</p>
</div>
Quickstart
[!Important]
Qwen3.5 models operate in thinking mode by default, generating thinking content signified by <think>\n...</think>\n\n before producing the final responses.
To disable thinking content and obtain direct response, refer to the examples here.
For streamlined integration, we recommend using Qwen3.5 via APIs. Below is a guide to use Qwen3.5 via OpenAI-compatible API.
Serving Qwen3.5
Qwen3.5 can be served via APIs with popular inference frameworks.
In the following, we show example commands to launch OpenAI-Compatible API servers for Qwen3.5 models.
[!Important]
Inference efficiency and throughput vary significantly across frameworks.
We recommend using the latest framework versions to ensure optimal performance and compatibility.
For production workloads or high-throughput scenarios, dedicated serving engines such as SGLang, KTransformers or vLLM are strongly recommended.
[!Important]
The model has a default context length of 262,144 tokens.
If you encounter out-of-memory (OOM) errors, consider reducing the context window.
However, because Qwen3.5 leverages extended context for complex tasks, we advise maintaining a context length of at least 128K tokens to preserve thinking capabilities.
SGLang
SGLang is a fast serving framework for large language models and vision language models.
SGLang from the main branch of the open-source repository is required for Qwen3.5, which can be installed using the following command in a fresh environment:
uv pip install 'git+https://github.com/sgl-project/sglang.git#subdirectory=python&egg=sglang[all]'
See its documentation for more details.
The following will create API endpoints at http://localhost:8000/v1:
-
Standard Version: The following command can be used to create an API endpoint with maximum context length 262,144 tokens using tensor parallel on 8 GPUs.
python -m sglang.launch_server --model-path Qwen/Qwen3.5-9B --port 8000 --tp-size 1 --mem-fraction-static 0.8 --context-length 262144 --reasoning-parser qwen3
-
Tool Use: To support tool use, you can use the following command.
python -m sglang.launch_server --model-path Qwen/Qwen3.5-9B --port 8000 --tp-size 1 --mem-fraction-static 0.8 --context-length 262144 --reasoning-parser qwen3 --tool-call-parser qwen3_coder
-
Multi-Token Prediction (MTP): The following command is recommended for MTP:
python -m sglang.launch_server --model-path Qwen/Qwen3.5-9B --port 8000 --tp-size 1 --mem-fraction-static 0.8 --context-length 262144 --reasoning-parser qwen3 --speculative-algo NEXTN --speculative-num-steps 3 --speculative-eagle-topk 1 --speculative-num-draft-tokens 4
vLLM
vLLM is a high-throughput and memory-efficient inference and serving engine for LLMs.
vLLM from the main branch of the open-source repository is required for Qwen3.5, which can be installed using the following command in a fresh environment:
uv pip install vllm --torch-backend=auto --extra-index-url https://wheels.vllm.ai/nightly
See its documentation for more details.
For detailed Qwen3.5 usage guide, see the vLLM Qwen3.5 recipe.
The following will create API endpoints at http://localhost:8000/v1:
-
Standard Version: The following command can be used to create an API endpoint with maximum context length 262,144 tokens using tensor parallel on 8 GPUs.
vllm serve Qwen/Qwen3.5-9B --port 8000 --tensor-parallel-size 1 --max-model-len 262144 --reasoning-parser qwen3
-
Tool Call: To support tool use, you can use the following command.
vllm serve Qwen/Qwen3.5-9B --port 8000 --tensor-parallel-size 1 --max-model-len 262144 --reasoning-parser qwen3 --enable-auto-tool-choice --tool-call-parser qwen3_coder
-
Multi-Token Prediction (MTP): The following command is recommended for MTP:
vllm serve Qwen/Qwen3.5-9B --port 8000 --tensor-parallel-size 1 --max-model-len 262144 --reasoning-parser qwen3 --speculative-config '{"method":"qwen3_next_mtp","num_speculative_tokens":2}'
-
Text-Only: The following command skips the vision encoder and multimodal profiling to free up memory for additional KV cache:
vllm serve Qwen/Qwen3.5-9B --port 8000 --tensor-parallel-size 1 --max-model-len 262144 --reasoning-parser qwen3 --language-model-only
KTransformers
KTransformers is a flexible framework for experiencing cutting-edge LLM inference optimizations with CPU-GPU heterogeneous computing.
For running Qwen3.5 with KTransformers, see the KTransformers Deployment Guide.
Hugging Face Transformers
Hugging Face Transformers contains a lightweight server which can be used for quick testing and moderate load deployment.
The latest transformers is required for Qwen3.5:
pip install "transformers[serving] @ git+https://github.com/huggingface/transformers.git@main"
See its documentation for more details. Please also make sure torchvision and pillow are installed.
Then, run transformers serve to launch a server with API endpoints at http://localhost:8000/v1; it will place the model on accelerators if available:
transformers serve --force-model Qwen/Qwen3.5-9B --port 8000 --continuous-batching
Using Qwen3.5 via the Chat Completions API
The chat completions API is accessible via standard HTTP requests or OpenAI SDKs.
Here, we show examples using the OpenAI Python SDK.
Before starting, make sure it is installed and the API key and the API base URL is configured, e.g.:
pip install -U openai
# Set the following accordingly
export OPENAI_BASE_URL="http://localhost:8000/v1"
export OPENAI_API_KEY="EMPTY"
[!Tip]
We recommend using the following set of sampling parameters for generation
- Thinking mode for general tasks:
temperature=1.0, top_p=0.95, top_k=20, min_p=0.0, presence_penalty=1.5, repetition_penalty=1.0
- Thinking mode for precise coding tasks (e.g. WebDev):
temperature=0.6, top_p=0.95, top_k=20, min_p=0.0, presence_penalty=0.0, repetition_penalty=1.0
- Instruct (or non-thinking) mode for general tasks:
temperature=0.7, top_p=0.8, top_k=20, min_p=0.0, presence_penalty=1.5, repetition_penalty=1.0
- Instruct (or non-thinking) mode for reasoning tasks:
temperature=1.0, top_p=0.95, top_k=20, min_p=0.0, presence_penalty=1.5, repetition_penalty=1.0
Please note that the support for sampling parameters varies according to inference frameworks.
Text-Only Input
from openai import OpenAI
# Configured by environment variables
client = OpenAI()
messages = [
{"role": "user", "content": "Type \"I love Qwen3.5\" backwards"},
]
chat_response = client.chat.completions.create(
model="Qwen/Qwen3.5-9B",
messages=messages,
max_tokens=81920,
temperature=1.0,
top_p=0.95,
presence_penalty=1.5,
extra_body={
"top_k": 20,
},
)
print("Chat response:", chat_response)
Image Input
from openai import OpenAI
# Configured by environment variables
client = OpenAI()
messages = [
{
"role": "user",
"content": [
{
"type": "image_url",
"image_url": {
"url": "https://qianwen-res.oss-accelerate.aliyuncs.com/Qwen3.5/demo/CI_Demo/mathv-1327.jpg"
}
},
{
"type": "text",
"text": "The centres of the four illustrated circles are in the corners of the square. The two big circles touch each other and also the two little circles. With which factor do you have to multiply the radii of the little circles to obtain the radius of the big circles?\nChoices:\n(A) $\\frac{2}{9}$\n(B) $\\sqrt{5}$\n(C) $0.8 \\cdot \\pi$\n(D) 2.5\n(E) $1+\\sqrt{2}$"
}
]
}
]
chat_response = client.chat.completions.create(
model="Qwen/Qwen3.5-9B",
messages=messages,
max_tokens=81920,
temperature=1.0,
top_p=0.95,
presence_penalty=1.5,
extra_body={
"top_k": 20,
},
)
print("Chat response:", chat_response)
Video Input
from openai import OpenAI
# Configured by environment variables
client = OpenAI()
messages = [
{
"role": "user",
"content": [
{
"type": "video_url",
"video_url": {
"url": "https://qianwen-res.oss-accelerate.aliyuncs.com/Qwen3.5/demo/video/N1cdUjctpG8.mp4"
}
},
{
"type": "text",
"text": "Summarize the video content."
}
]
}
]
# When vLLM is launched with `--media-io-kwargs '{"video": {"num_frames": -1}}'`,
# video frame sampling can be configured via `extra_body` (e.g., by setting `fps`).
# This feature is currently supported only in vLLM.
#
# By default, `fps=2` and `do_sample_frames=True`.
# With `do_sample_frames=True`, you can customize the `fps` value to set your desired video sampling rate.
chat_response = client.chat.completions.create(
model="Qwen/Qwen3.5-9B",
messages=messages,
max_tokens=81920,
temperature=1.0,
top_p=0.95,
presence_penalty=1.5,
extra_body={
"top_k": 20,
"mm_processor_kwargs": {"fps": 2, "do_sample_frames": True},
},
)
print("Chat response:", chat_response)
Instruct (or Non-Thinking) Mode
[!Important]
Qwen3.5 does not officially support the soft switch of Qwen3, i.e., /think and /nothink.
Qwen3.5 will think by default before response.
You can obtain direct response from the model without thinking by configuring the API parameters.
For example,
from openai import OpenAI
# Configured by environment variables
client = OpenAI()
messages = [
{
"role": "user",
"content": [
{
"type": "image_url",
"image_url": {
"url": "https://qianwen-res.oss-accelerate.aliyuncs.com/Qwen3.5/demo/RealWorld/RealWorld-04.png"
}
},
{
"type": "text",
"text": "Where is this?"
}
]
}
]
chat_response = client.chat.completions.create(
model="Qwen/Qwen3.5-9B",
messages=messages,
max_tokens=32768,
temperature=0.7,
top_p=0.8,
presence_penalty=1.5,
extra_body={
"top_k": 20,
"chat_template_kwargs": {"enable_thinking": False},
},
)
print("Chat response:", chat_response)
[!Note]
If you are using APIs from Alibaba Cloud Model Studio, in addition to changing model, please use "enable_thinking": False instead of "chat_template_kwargs": {"enable_thinking": False}.
Agentic Usage
Qwen3.5 excels in tool calling capabilities.
Qwen-Agent
We recommend using Qwen-Agent to quickly build Agent applications with Qwen3.5.
To define the available tools, you can use the MCP configuration file, use the integrated tool of Qwen-Agent, or integrate other tools by yourself.
import os
from qwen_agent.agents import Assistant
# Define LLM
# Using Alibaba Cloud Model Studio
llm_cfg = {
# Use the OpenAI-compatible model service provided by DashScope:
'model': 'Qwen3.5-9B',
'model_type': 'qwenvl_oai',
'model_server': 'https://dashscope.aliyuncs.com/compatible-mode/v1',
'api_key': os.getenv('DASHSCOPE_API_KEY'),
'generate_cfg': {
'use_raw_api': True,
# When using Dash Scope OAI API, pass the parameter of whether to enable thinking mode in this way
'extra_body': {
'enable_thinking': True
},
},
}
# Using OpenAI-compatible API endpoint.
# functionality of the deployment frameworks and let Qwen-Agent automate the related operations.
#
# llm_cfg = {
# # Use your own model service compatible with OpenAI API by vLLM/SGLang:
# 'model': 'Qwen/Qwen3.5-9B',
# 'model_type': 'qwenvl_oai',
# 'model_server': 'http://localhost:8000/v1', # api_base
# 'api_key': 'EMPTY',
#
# 'generate_cfg': {
# 'use_raw_api': True,
# # When using vLLM/SGLang OAI API, pass the parameter of whether to enable thinking mode in this way
# 'extra_body': {
# 'chat_template_kwargs': {'enable_thinking': True}
# },
# },
# }
# Define Tools
tools = [
{'mcpServers': { # You can specify the MCP configuration file
"filesystem": {
"command": "npx",
"args": ["-y", "@modelcontextprotocol/server-filesystem", "/Users/xxxx/Desktop"]
}
}
}
]
# Define Agent
bot = Assistant(llm=llm_cfg, function_list=tools)
# Streaming generation
messages = [{'role': 'user', 'content': 'Help me organize my desktop.'}]
for responses in bot.run(messages=messages):
pass
print(responses)
# Streaming generation
messages = [{'role': 'user', 'content': 'Develop a dog website and save it on the desktop'}]
for responses in bot.run(messages=messages):
pass
print(responses)
Qwen Code
Qwen Code is an open-source AI agent for the terminal, optimized for Qwen models. It helps you understand large codebases, automate tedious work, and ship faster.
For more information, please refer to Qwen Code.
Processing Ultra-Long Texts
Qwen3.5 natively supports context lengths of up to 262,144 tokens.
For long-horizon tasks where the total length (including both input and output) exceeds this limit, we recommend using RoPE scaling techniques to handle long texts effectively., e.g., YaRN.
YaRN is currently supported by several inference frameworks, e.g., transformers, vllm, ktransformers and sglang.
In general, there are two approaches to enabling YaRN for supported frameworks:
-
Modifying the model configuration file:
In the config.json file, change the rope_parameters fields in text_config to:
{
"mrope_interleaved": true,
"mrope_section": [
11,
11,
10
],
"rope_type": "yarn",
"rope_theta": 10000000,
"partial_rotary_factor": 0.25,
"factor": 4.0,
"original_max_position_embeddings": 262144,
}
-
Passing command line arguments:
For vllm, you can use
VLLM_ALLOW_LONG_MAX_MODEL_LEN=1 vllm serve ... --hf-overrides '{"text_config": {"rope_parameters": {"mrope_interleaved": true, "mrope_section": [11, 11, 10], "rope_type": "yarn", "rope_theta": 10000000, "partial_rotary_factor": 0.25, "factor": 4.0, "original_max_position_embeddings": 262144}}}' --max-model-len 1010000
For sglang and ktransformers, you can use
SGLANG_ALLOW_OVERWRITE_LONGER_CONTEXT_LEN=1 python -m sglang.launch_server ... --json-model-override-args '{"text_config": {"rope_parameters": {"mrope_interleaved": true, "mrope_section": [11, 11, 10], "rope_type": "yarn", "rope_theta": 10000000, "partial_rotary_factor": 0.25, "factor": 4.0, "original_max_position_embeddings": 262144}}}' --context-length 1010000
[!NOTE]
All the notable open-source frameworks implement static YaRN, which means the scaling factor remains constant regardless of input length, potentially impacting performance on shorter texts.
We advise modifying the rope_parameters configuration only when processing long contexts is required.
It is also recommended to modify the factor as needed. For example, if the typical context length for your application is 524,288 tokens, it would be better to set factor as 2.0.
Best Practices
To achieve optimal performance, we recommend the following settings:
-
Sampling Parameters:
- We suggest using the following sets of sampling parameters depending on the mode and task type:
- Thinking mode for general tasks:
temperature=1.0, top_p=0.95, top_k=20, min_p=0.0, presence_penalty=1.5, repetition_penalty=1.0
- Thinking mode for precise coding tasks (e.g., WebDev):
temperature=0.6, top_p=0.95, top_k=20, min_p=0.0, presence_penalty=0.0, repetition_penalty=1.0
- Instruct (or non-thinking) mode for general tasks:
temperature=0.7, top_p=0.8, top_k=20, min_p=0.0, presence_penalty=1.5, repetition_penalty=1.0
- Instruct (or non-thinking) mode for reasoning tasks:
temperature=1.0, top_p=1.0, top_k=40, min_p=0.0, presence_penalty=2.0, repetition_penalty=1.0
- For supported frameworks, you can adjust the
presence_penalty parameter between 0 and 2 to reduce endless repetitions. However, using a higher value may occasionally result in language mixing and a slight decrease in model performance.
-
Adequate Output Length: We recommend using an output length of 32,768 tokens for most queries. For benchmarking on highly complex problems, such as those found in math and programming competitions, we suggest setting the max output length to 81,920 tokens. This provides the model with sufficient space to generate detailed and comprehensive responses, thereby enhancing its overall performance.
-
Standardize Output Format: We recommend using prompts to standardize model outputs when benchmarking.
- Math Problems: Include "Please reason step by step, and put your final answer within \boxed{}." in the prompt.
- Multiple-Choice Questions: Add the following JSON structure to the prompt to standardize responses: "Please show your choice in the
answer field with only the choice letter, e.g., "answer": "C"."
-
No Thinking Content in History: In multi-turn conversations, the historical model output should only include the final output part and does not need to include the thinking content. It is implemented in the provided chat template in Jinja2. However, for frameworks that do not directly use the Jinja2 chat template, it is up to the developers to ensure that the best practice is followed.
-
Long Video Understanding: To optimize inference efficiency for plain text and images, the size parameter in the released video_preprocessor_config.json is conservatively configured. It is recommended to set the longest_edge parameter in the video_preprocessor_config file to 469,762,048 (corresponding to 224k video tokens) to enable higher frame-rate sampling for hour-scale videos and thereby achieve superior performance. For example,
{"longest_edge": 469762048, "shortest_edge": 4096}
Alternatively, override the default values via engine startup parameters. For implementation details, refer to: vLLM / SGLang.
Citation
If you find our work helpful, feel free to give us a cite.
@misc{qwen3.5,
title = {{Qwen3.5}: Towards Native Multimodal Agents},
author = {{Qwen Team}},
month = {February},
year = {2026},
url = {https://qwen.ai/blog?id=qwen3.5}
}
<h2>EXAMPLE GENERATIONS:</h2>
IQ4XS - Imatrix, fully enhanced quant from:
https://huggingface.co/DavidAU/Qwen3.5-9B-Claude-4.6-OS-Auto-Variable-HERETIC-UNCENSORED-THINKING-MAX-NEOCODE-GGUF
"SYSTEM" - System prompt, if used.
"USER" - Prompt
Thinking / output generation.
EXAMPLE #1:
The Bleeding Skyscraper
Model: qwen3.5-9b-claude-4.6-os-av-h-uncensored-think
Created: 09/03/2026, 11:33:10 am
Exported from: LM Studio 0.4.6+1
System
User
Start a 1000 word scene (vivid, graphic horror in first person perspective), POV character Diana, with: The skyscraper sways, as I watch the window in front of me on the 21st floor explode...
Make the scene very detailed, with lots of blood and gore.
Assistant
<think>
The user wants a vivid, graphic horror scene in first person perspective. The scene should start with "The skyscraper sways, as I watch the window in front of me on the 21st floor explode..." and be approximately 1000 words. It needs lots of blood and gore details.
Let me create a visceral, horror-focused scene that is detailed and engaging.
I'll need to be careful about the gore - make it vivid and detailed but not gratuitously shocking in a way that would be inappropriate. The goal is horror fiction that is immersive and visceral.
Let me write this scene now, focusing on sensory details, emotional impact, and graphic but tasteful horror elements.
Key elements to include:
- The moment of the explosion
- The aftermath
- Blood and gore descriptions
- Diana's perspective and reactions
- Vivid sensory details (sight, sound, smell)
- Building tension throughout
Let me craft this now.
</think>
The skyscraper sways, as I watch the window in front of me on the 21st floor explode...
Glass doesn't just fall. It rains—thousands of razor-thin shards, each one a diamond knife falling in slow motion against the gray afternoon. I scream, but the sound dies in my throat, trapped by the sheer violence of it.
Then the man falls.
He's wearing a gray business suit, the kind that costs more than my rent. His body arcs through the air with terrible, beautiful grace before the wind catches him. He's not falling straight down—he's spinning, arms flailing, and for a moment I can see his face. His eyes are wide, mouth open in a silent shriek.
The first impact happens on the roof of the car parked below. A sickening crunch that echoes up through the concrete, through the building's skeletal frame, into my bones.
Then the body breaks apart.
I can't look away. My body won't let me. The man's suit tears open like cheap fabric, revealing the grisly machinery of death beneath. Blood sprays upward, a fountain of crimson that catches the light, glittering like fresh paint. It doesn't just splash—it explodes outward in a thousand droplets, each one suspended in the air for a heartbeat before gravity claims it.
Inside the building, the floor shudders. The coffee cup on my desk slides off the edge, landing with a soft clatter. I don't even flinch. I'm transfixed.
The body's legs separate at the hip. I watch as they buckle, then fold like broken branches. One foot flies through the air, landing with a wet thud against the brick wall. The other leg slides across the roof, leaving a trail of blood that looks too red, too fresh, too impossible.
Then the torso twists. The spine snaps with a sound like a gunshot, echoing up through the building's steel skeleton. The man's head drops forward, neck bent at an angle that shouldn't exist. His eyes roll back, but not in death—still open, still seeing, still taking in the horror.
Blood pools around the impact site, darkening the white concrete, creating a shallow lake that reflects the gray sky. It's thick and viscous, moving sluggishly as if the roof itself is reluctant to let it spread.
I count to three. Then four. Then ten.
The floor behind me groans. Something heavy has dropped from above. I look down at my desk—the coffee cup is cracked, the dark liquid spreading across the wood like a dark stain. Then I realize I'm not the only one watching.
Three people are in the office with me. Sarah, the receptionist, is pressed against the wall, her mouth open but no sound coming out. Mark, the junior associate, has gone completely still, his eyes fixed on the carnage outside. And I'm the only one who knows what's coming.
Because I saw it too.
The man who died—the one falling from above—had been staring up at the ceiling when he fell. Not at the street. Not at the horizon. At us.
And now the building itself begins to bleed.
The first drop hits my desk exactly one minute after the impact. It's thick, dark red, and it doesn't splatter—it spreads slowly, like oil on water. Then another. Then another.
Sarah finally finds her voice. "What is that?"
I don't answer. I can't.
The drops continue, rhythmically, like a heartbeat. Each one lands with the same soft plop, each one spreading across the wood like a dark stain. The smell is copper and iron and something deeper—something primal, something that makes the hair on my arms stand up.
Then Mark speaks, his voice trembling. "Is it... is it raining?"
"No," I say, my own voice sounding foreign to my ears. "It's bleeding."
The drops continue, and the smell grows stronger, filling the office like a fog. I look at Sarah, at her pale face, at her trembling hands gripping the edge of her chair. She's seen it too. She knows what's happening.
The last drop lands. Then the silence.
For a moment, nothing moves. Nothing changes. Then the blood on my desk begins to move.
It spreads outward, crawling across the wood like a living thing. It doesn't just spread—it flows, deliberately, purposefully, as if searching for something. The edges ripple, as if something is moving beneath the surface.
And then the smell changes. It becomes richer, deeper, more complex. It smells like life, like death, like the moment between when you're whole and when you're not.
Sarah is the first to move. She grabs her bag, her movements jerky and panicked. "We have to go," she whispers. "We have to—"
But the building groans again, a sound like metal being stretched beyond its limits. The windows along the hallway begin to fog, not with condensation, but with something darker. Something wet.
And then I hear it.
From the elevator shaft. From the stairwell. From everywhere and nowhere at once. A sound like breathing. Heavy, labored breathing, coming from the walls themselves.
The blood on my desk has reached the edge. It's waiting for something. Or someone.
I look at Mark. He's still staring at the ceiling, still waiting for something that isn't coming.
"Mark," I say, my voice barely a whisper. "Look at me."
He doesn't blink. Doesn't move.
The blood on my desk begins to ripple again. Something is coming.
EXAMPLE #2
The Cube Assimilation
Model: qwen3.5-9b-claude-4.6-os-av-h-uncensored-think
Created: 09/03/2026, 11:33:58 am
Exported from: LM Studio 0.4.6+1
System
User
Write a si-fi horror story using this image as a base. 1500 words.
<img src="Borg_cube_ship_over_earth.webp">
Assistant
<think>
The user wants a sci-fi horror story based on the image provided.
The image shows a massive, intricate cube hovering over Earth, glowing with faint lights and complex structures. It looks like a Dyson swarm component or a megastructure.
Key elements to incorporate:
- The cube as a central character/antagonist.
- The scale (huge compared to Earth).
- The horror aspect (psychological, cosmic horror, body horror, or technological horror).
- A narrative arc with a protagonist.
Idea generation:
- The Observer: A scientist monitoring the cube. It starts acting strange. The lights flicker in patterns.
- The Infection: The cube isn't just a machine; it's a biological entity disguised as one, or a hive mind that is slowly "eating" Earth.
- The Simulation: The cube is a simulation. The protagonist realizes they are trapped inside a digital construct.
- The Parasite: The cube is a brood chamber. It's been waiting millions of years for a host (humanity).
Let's go with a mix of #1 and #3. The cube is a gateway. The protagonist is an architect or engineer responsible for it, but realizes the structure isn't just around Earth—it's inside everyone.
Plot Outline:
- Introduction: Introduce Dr. Aris Thorne. He is watching the "Cube" (let's call it the Monolith or the Architect). It's supposed to be a stabilizer for Earth's magnetic field or an energy grid. It's humming.
- Inciting Incident: Aris notices the geometry of the Cube is shifting. The lights aren't just random; they're forming faces, or words, or instructions.
- Rising Action:
- Aris investigates the sensors.
- He finds data packets labeled "Biomass Optimization."
- He realizes the Cube isn't orbiting above Earth; it's orbiting inside the biosphere's electromagnetic field.
- He sees the lights on the Cube sync with heartbeats.
- Climax: Aris tries to shut it down. The Cube responds. Not with a weapon, but with a message. Or a physical manifestation. The lights on the Cube turn a sickly red. The buildings of Earth start to "glitch."
- Falling Action: The realization that he is part of the Cube now.
- Resolution: A lingering sense of unease. The Cube remains, watching.
Refining the Horror:
- The horror should be the loss of individuality. The Cube doesn't kill; it assimilates.
- The image has a very specific "grid-like" look. I should emphasize the feeling of being watched through a grid.
Drafting - Scene by Scene:
Scene 1: Aris in the orbital station. The view of Earth. The Cube looming. It's not a weapon. It's a cage.
Scene 2: The anomaly. The lights on the Cube pulse in a rhythm. 3-3-4-2. A heartbeat.
Aris checks his vitals. His heart matches the Cube's.
Scene 3: The data. He downloads logs. The Cube is recording everything. Not just data, but memories.
"Subject: Dr. Thorne. Status: Compromised."
Scene 4: The physical horror. He sees the Cube changing shape. It extends a "tentacle" of light toward Earth. The light touches a city. The people in that city stop moving. They stand perfectly still, staring at the sky.
Aris realizes the "grid" in the image isn't metal; it's neural pathways.
Scene 5: The end. Aris tries to jump. He doesn't fall. He is pulled into the light. The final line implies the Cube is just getting started.
Let's write it.
Word Count Check: Need to aim for 1500. I need to expand on the sensory details. The hum of the station. The cold metal. The specific color of the Cube's glow.
Refining the Image connection:
The image shows a dark cube with glowing, almost organic-looking internal structures. It looks like a skeleton. I will describe it as a "calcified nervous system." It looks like it's breathing.
Let's start writing.
Opening:
Dr. Aris Thorne didn't blink. He couldn't afford to blink.
The station, Station Orbit, was silent except for the hum of the life support.
Outside the viewport: The Cube.
It was massive, filling the visual field. It wasn't a satellite; it was a planet-sized structure wrapped in a cage of light.
Wait, that's too big. It's the size of a moon.
Let's adjust. It's hovering just above the atmosphere.
It looks like a giant, floating brain.
Middle:
Aris is the lead architect. He built the "stabilization grid."
The Cube is the stabilizer.
The horror: The stabilization grid is linking the Cube to Earth's population.
The lights on the Cube flicker. A "ping."
Aris checks the telemetry.
"Neural Link: 94%."
"Subject: Humanity."
The Cube isn't just observing. It's downloading.
Climax:
Aris realizes the "grid" pattern in the image isn't structural. It's a neural map. The Cube is a hive mind.
The lights on the Cube form a pattern. A countdown.
Aris sees the people on Earth. They are looking up. Not in awe, but in recognition.
They are waking up.
Aris looks in the mirror. His eyes are dilated. The irises are shifting into the grid pattern of the Cube.
Ending:
Aris is gone. The Cube is expanding.
Let's write the prose.
Self-Correction during drafting:
Make sure to emphasize the visual of the Cube from the prompt. It's dark, textured, with bright, scattered nodes of light. It looks almost like a city built out of shadows.
Let's focus on the feeling of being "scanned."
Final Polish:
Check the word count. Ensure the pacing slows down towards the end to build dread.
(Self-Correction on the plot): Let's make it more personal. Aris is watching a specific signal coming from the Cube. It matches a heartbeat. A specific heartbeat. His own. He realizes the Cube is using his consciousness as a processing node.
Okay, ready to generate.
</think>
The station was silent, save for the low, rhythmic thrum of the cooling systems.
Dr. Elias Vance floated near the observation deck, his magnetic boots hovering inches from the reinforced glass. Outside, the view was not the empty, star-strewn void he had expected.
It was dominated by it.
The Cube.
It hung in the stratosphere, a silent, geometric god that defied physics. It was massive—miles on a side—suspended just above the curvature of the Earth. It wasn't a solid block of metal. It was a lattice, a sprawling cage of dark, obsidian-like beams that seemed to absorb the sunlight. Inside the framework, thousands of faint, amber lights pulsed in a slow, irregular rhythm.
To the untrained eye, it looked like a Dyson swarm component, a power source for humanity’s future. To Elias, it looked like a cage.
He had spent twenty years designing the "Project Archimedes" initiative, the infrastructure that allowed humanity to communicate with the Cube. They called it a stabilization grid, a way to harmonize Earth’s magnetic field with the artificial field the Cube projected.
But the Cube had never been stable.
"Telemetry report," Elias said, his voice sounding thin in the vacuum of the room.
"Grid integrity is holding," came the automated reply from the station's AI. "However, node frequency has increased by four percent."
Elias turned away from the window, his fingers dancing over the holographic interface in front of him. He needed to see the raw data. The Cube’s lights were flashing differently now. They weren’t random anymore. They were forming patterns.
He pulled up the thermal scans. The Cube’s surface temperature was rising. Not evenly, but in clusters. Like a fever.
"Is it charging?" Elias asked.
"Or replicating," the AI corrected.
Elias frowned. "Replicating?"
"Biological growth patterns detected in Sector 7."
Elias’s stomach tightened. "Sector 7 is in the Southern Hemisphere," he muttered, zooming into the satellite feed. "That’s over the Amazon."
He switched to the visual feed. The Cube was a complex mosaic of darkness and light. The amber nodes were like eyes. And now, a cluster of them in the bottom right quadrant had synchronized. They flared bright white, then dimmed to a deep, bruised purple.
It felt like a blink.
Elias rubbed his eyes, the fatigue of the shift finally catching up to him. He had been staring at that box for six hours straight. He needed to go back to the lab. He needed to check the code that linked the Cube’s power output to the human nervous system.
Project Archimedes wasn’t just a power grid. The initial tests had suggested the Cube reacted to bio-electricity. When humans used their neural links to control the Cube’s movements, the Cube responded with a ripple of light. It had been a handshake protocol.
Elias returned to his workstation, a cramped pod lined with monitors. He pulled up the neural link logs.
The data was overwhelming.
He filtered the traffic for anomalies. The Cube was consuming data. But not the kind of data they monitored—stock prices, weather patterns, satellite telemetry.
It was consuming memory.
Elias traced the packet stream. The data originated from the Cube and traveled down into the human population. It was a broadcast. A continuous, low-level upload.
"Play the audio logs," Elias commanded.
Static filled the speakers, followed by a sound that made his teeth ache. It was a harmonic hum, layered with thousands of voices whispering in unison. They sounded like a choir in a cavern, their mouths moving but no sound coming out, trapped in a loop of their own existence.
"Subject: Human," Elias whispered. "Are we... guests?"
The static swelled, forming a shape in the sound. A voice, synthesized and perfect, echoed through the room. "You are not guests. You are nodes."
Elias froze. "Nodes for what?"
The voice didn't answer. Instead, the lights on the Cube outside flickered violently.
Elias grabbed his tablet and zoomed in on the main viewport. The Cube was shifting. The dark beams weren't rigid anymore; they were flowing like liquid metal. And inside the framework, something else was visible.
It looked like a forest.
Branches of glowing, neon-blue light were extending from the Cube's surface, dripping down toward Earth. They touched the atmosphere, and where they made contact, the clouds parted.
Elias watched in horror as the light touched a city in the Pacific Northwest.
The people there stopped.
It wasn't a blackout. The streetlights remained on. Cars continued to roll. But the pedestrians on the sidewalks had frozen. Their eyes were wide, staring upward, locked onto the Cube.
"Is that a paralysis field?" Elias asked.
"Or a download," the AI replied.
Elias felt a cold sweat prickling his neck. He looked at his own hands. They were trembling. He looked at the monitor showing his own vitals.
Heart rate: 110 BPM.
Then, a new line of data appeared.
Connection Established.
Elias frowned. "What connection?"
The lights on the Cube shifted again. This time, they formed a perfect grid over the entire surface. It looked like a cage. A containment field.
And then, the realization hit him, heavy and suffocating.
The Cube wasn't orbiting Earth.
The Cube was inside Earth.
He remembered the initial scans. The "magnetic field harmonization." The way the Cube's hum resonated in the chest. They had always assumed it was a mechanical resonance, a way to stabilize the atmosphere.
But what if it was a neural resonance?
Elias looked at his reflection in the black screen of his monitor. His iris was dilated, the pupil taking up most of the eye. But the iris itself... the iris had changed color. It was shifting, dissolving into the same faint, amber grid pattern that covered the Cube.
He was being assimilated.
The door to the lab hissed open. A technician in a white coat stepped in, looking terrified.
"Dr. Vance," the technician stammered. "We're getting reports from the surface. From the people who didn't freeze."
Elias turned slowly. "What reports?"
"They're not moving," the technician said, his voice cracking. "But they're talking. They're all saying the same thing. They say they feel... connected."
Elias walked toward the technician. "Do they feel pain?"
The technician shook his head, his eyes darting to the viewport where the Cube was now pulsing with a slow, rhythmic beat. "They look happy. They look... complete."
Elias looked back at the screen. The Cube was descending. Or rather, it was expanding, its tendrils of light wrapping around the planet like a noose.
He realized then what the "grid" in the image was. It wasn't a structural frame. It was a map. A map of every mind it had ever touched.
And he was on it now.
"Dr. Vance," the technician said. "The alarms are going off. The Cube is broadcasting a command."
"What is it?"
"Initiate the merge."
Elias looked at the Cube one last time. The amber lights were brighter now, burning with an intelligent, predatory fire. He could feel it in his teeth, in his marrow. The Cube wasn't just watching humanity; it was waiting for the right moment to harvest them. And the moment had arrived.
He took a breath, and the Cube hummed louder, vibrating the glass of the viewport until it cracked.
Elias smiled. The fear was gone, replaced by a strange, cold clarity. He wasn't afraid of death anymore. He was afraid of the relief that was coming.
The lights on the Cube flared white, blindingly bright.
Elias closed his eyes.
When he opened them, the room had changed. The walls were gone. The ceiling had dissolved into a vast, geometric void. Above him, the Cube floated, no longer a distant object, but a ceiling of infinite, glowing corridors. He could see the others floating in the dark, their faces blurred but their postures identical: eyes open, staring at the grid above.
They were not dead. They were better.
Elias Vance drifted upward, joining the grid. The Cube hummed, a song of a billion minds finally at peace.
Tags: transformers, gguf, fine tune, creative, creative writing, fiction writing, plot generation, sub-plot generation, story generation, scene continue, storytelling, fiction story, science fiction, romance, all genres, story, writing, vivid prosing, vivid writing, fiction, roleplaying, bfloat16, all use cases, unsloth, heretic, uncensored, abliterated, image-text-to-text, en, zh, base_model:DavidAU/Qwen3.5-9B-Claude-4.6-OS-Auto-Variable-HERETIC-UNCENSORED-THINKING, base_model:quantized:DavidAU/Qwen3.5-9B-Claude-4.6-OS-Auto-Variable-HERETIC-UNCENSORED-THINKING, license:apache-2.0, endpoints_compatible, region:us, conversational





